Note
Access to this page requires authorization. You can try signing in or changing directories.
Access to this page requires authorization. You can try changing directories.
Databricks Runtime supports the binary file data source, which reads binary files and converts each file into a single record that contains the raw content and metadata of the file. The binary file data source produces a DataFrame with the following columns and possibly partition columns:
path (StringType): The path of the file.modificationTime (TimestampType): The modification time of the file. In some Hadoop FileSystem implementations, this parameter might be unavailable and the value would be set to a default value.length (LongType): The length of the file in bytes.content (BinaryType): The contents of the file.
To read binary files, specify the data source format as binaryFile.
Images
Databricks recommends that you use the binary file data source to load image data.
The Databricks display function supports displaying image data loaded using the binary data source.
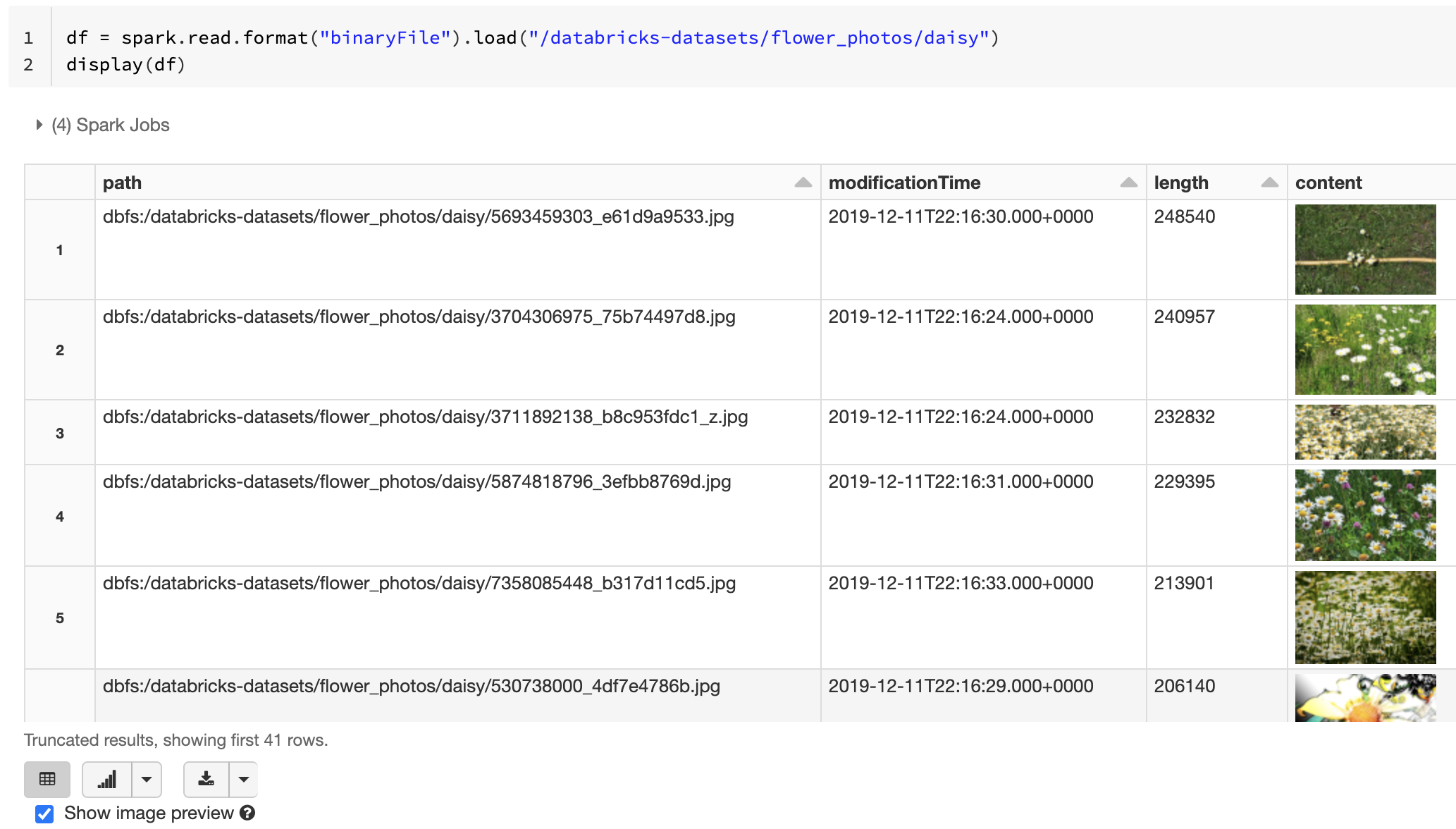
If all the loaded files have a file name with an image extension, image preview is automatically enabled:
df = spark.read.format("binaryFile").load("<path-to-image-dir>")
display(df) # image thumbnails are rendered in the "content" column
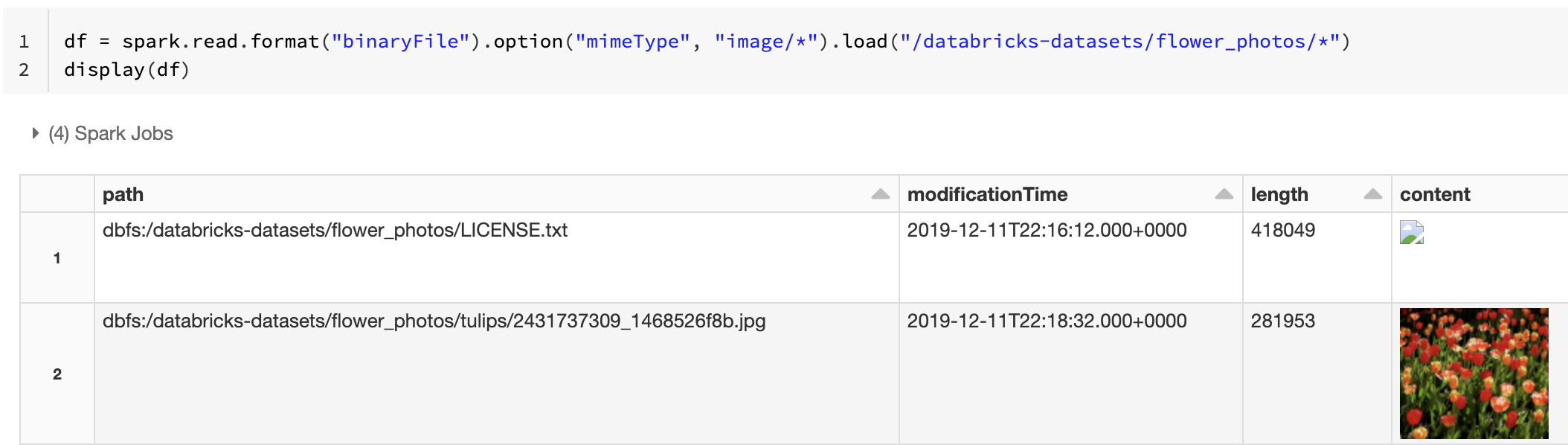
Alternatively, you can force the image preview functionality by using the mimeType option with a string value "image/*" to annotate the binary column. Images are decoded based on their format information in the binary content. Supported image types are bmp, gif, jpeg, and png. Unsupported files appear as a broken image icon.
df = spark.read.format("binaryFile").option("mimeType", "image/*").load("<path-to-dir>")
display(df) # unsupported files are displayed as a broken image icon
See Reference solution for image applications for the recommended workflow to handle image data.
Options
To load files with paths matching a given glob pattern while keeping the behavior of partition discovery,
you can use the pathGlobFilter option. The following code reads all JPG files from the
input directory with partition discovery:
df = spark.read.format("binaryFile").option("pathGlobFilter", "*.jpg").load("<path-to-dir>")
If you want to ignore partition discovery and recursively search files under the input directory,
use the recursiveFileLookup option. This option searches through nested directories
even if their names do not follow a partition naming scheme like date=2019-07-01.
The following code reads all JPG files recursively from the input directory and ignores partition discovery:
df = spark.read.format("binaryFile") \
.option("pathGlobFilter", "*.jpg") \
.option("recursiveFileLookup", "true") \
.load("<path-to-dir>")
Similar APIs exist for Scala, Java, and R.
Note
To improve read performance when you load data back, Azure Databricks recommends saving data loaded from binary files using Delta tables:
df.write.save("<path-to-table>")